Labelling cell components with neural networks
Problem description
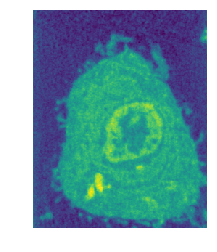
The National Center for Xray Tomography uses soft x-ray tomography to produce incredibly detailed images of whole cells. Their imaging techniques allow for high-resolution images of cells without staining or dehydrating, providing a detailed view of cells in their native state.
The images produced by the x-ray tomography are 3D arrays of densities (or linear absorption coefficients). Each array consists of approximately 400x400x400 voxels, each voxel a single floating point between 0 and 1. For many applications in cell biology and biomedical research, one wants accurate labellings of the various cell organelles. That is, we want to determine if a given voxel belongs to the cell cytoplasm, nucleus, mitochondria, etc. While it is possible for an expert to methodically label each of the 64 million voxel, even with the best existing software this is an extremely slow and laborious process. Therefore, any tools that can assist experts in quickly labelling the cellular data could provide enormous savings of time and energy, and allow for a vast increase in the amount of labelled data.
Methods
We propose a method based on recent advances in deep learning–a field which has shown striking success on the problems of image labelling and image segmentation. In particular, we use a convolutional neural network based on the architectures of DeconvNet and U-net to solve two binary classification problems: does a pixel belong to the cytoplasm; and, if so, does the pixel belong to the nucleus?
A human-expert labels the cell data one two-dimensional slice at a time. We employ a similar strategy, slicing the 3D array into 256x256 images. The output of our models is a 256x256 image of labels. Given our limited training data, we use a transfer-learning strategy. The architecture uses the first five convolution + max-pooling blocks of VGG-16, with weights trained on the ImageNet classification problem. These blocks are followed by five convolution + up-sampling blocks, along with skip through layers at blocks 2,3 and 4. We use Keras to implement and train the model.
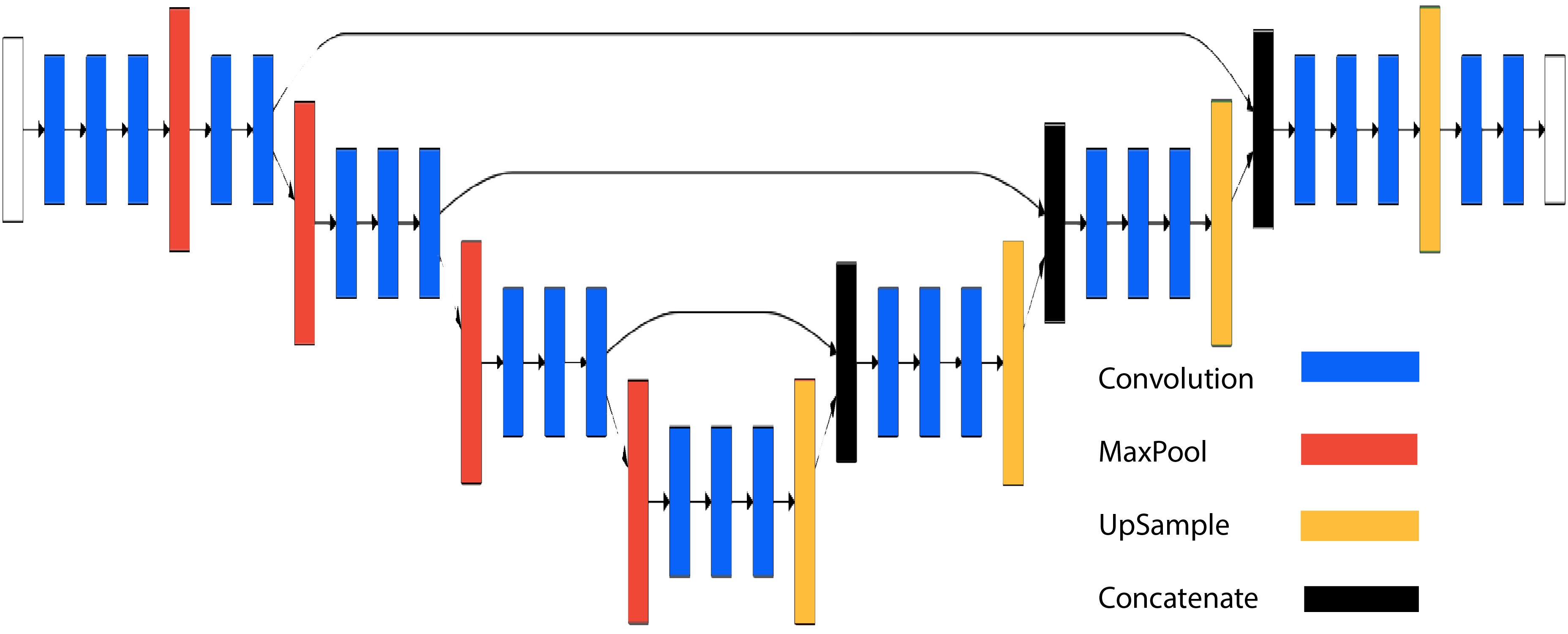
The deconvolutional layers of our model are trained on our limited data, and we find that the low-level features extracted by the VGG-16 layers work extremely well in our network: we achieve high accuracy with models trained on around 8000 training samples.
The three classes are extremely unbalanced: Only 3% of the voxels belong to the nucleus. We address the class imbalance by training two separate models: The first model determines if a voxel belongs to the cell or not. The second model determines if a voxel belongs to the nucleus. We oversample our training data for the second model so that a much higher proportion of training samples contain nucleus voxels.
Results
Cell Detection
Training on approximately 8000 samples, the cell detection model achieves an average accuracy of 0.9640 and an average Jaccard score of 0.8566 on cross-validation.
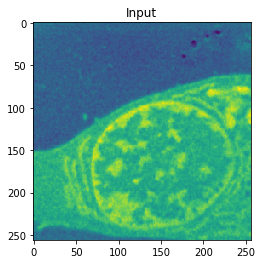
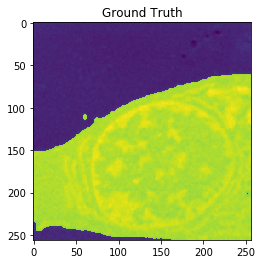
| Input | Ground truth | Predicted |
|---|---|---|
 |
 |
 |
| Accuracy: 0.9916 | Jaccard: 0.9851 | |
 |
 |
 |
| Accuracy: 0.9921 | Jaccard: 0.9870 | |
 |
 |
 |
| Accuracy: 0.9922 | Jaccard: 0.9824 |
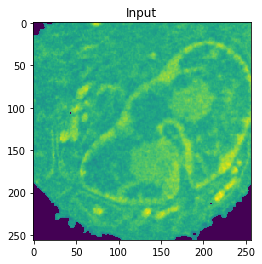
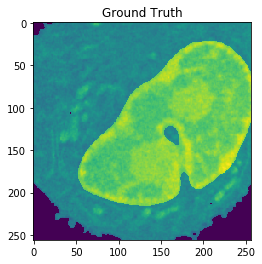
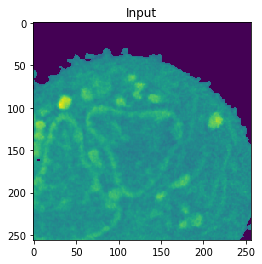
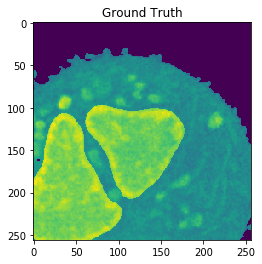
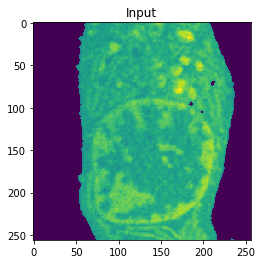
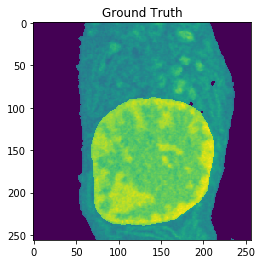
Nucleus Detection
Training on approximately 8000 samples, the nucleus model achieves an average accuracy 0.9784 and an average Jaccard score of 0.8642 on cross-validation.
| Input | Ground truth | Predicted |
|---|---|---|
 |
 |
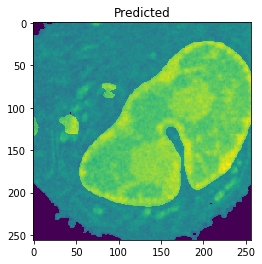 |
| Accuracy: 0.9802 | Jaccard: 0.9506 | |
 |
 |
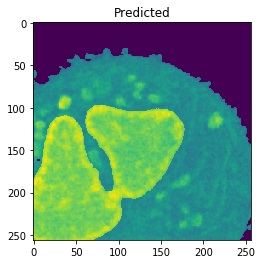 |
| Accuracy: 0.9866 | Jaccard: 0.9540 | |
 |
 |
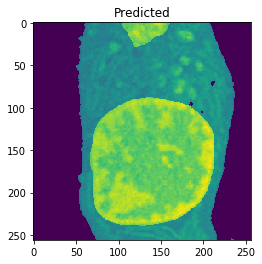 |
| Accuracy: 0.9767 | Jaccard: 0.9218 |
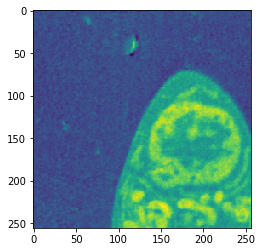
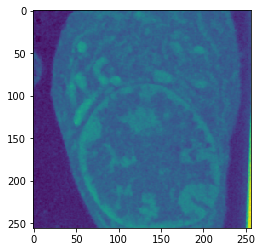
Combined
| Input | Ground truth | Predicted |
|---|---|---|
 |
 |
 |
 |
 |
 |
 |
 |
 |
Future work
These preliminary results suggest that variations of the U-net architecture, in tandem with transfer learning, can produce highly accurate segmentations of the cell images. There are many questions left to investigate–in particular:
- Can we achieve similar accuracies with simpler or shallower architectures?
- How does the model perform on different cell types? So far, the data is limited to human B-Cells.
- How much can we improve over-all accuracy by ensembling predictions on overlapping slices?
- We are currently predicting each slice independently. Can we combine these methods with LSTM architectures to more accurately label slices through 3D volumes?
Source
Code is available here.